Abstract
Breast cancer is the most common cancer in women worldwide. Current statistics show that one in eight women will develop this disease over the course of her lifetime. While X-ray mammography is the gold standard image modality for screening and diagnosis of breast cancer, it presents decreased sensitivity in dense breasts. Several studies have shown that the combination of the different medical image modalities, such as mammography and magnetic resonance imaging (MRI), leads to a more accurate diagnosis and, therefore, a more effective medical treatment of patient diseases. However, the fusion of information among several image modalities is a challenging task, due to the differences not only in the physics underlying each modality but, also, the different patient positioning during the image acquisition. The main purpose of this thesis is to evaluate the similarity among the information provided by two medical image modalities, such as the X-ray mammography and MRI, and, at the same time, to propose new algorithms to register the images in order to correlate the position of lesion and susceptible areas.
A deep review of the state-of-the-art, focusing our attention in the multimodal registration problems using patient-specific biomechanical finite-element (FE) models, is performed, from the biomechanical model construction (including the pre-processing and segmentation of MRI images, a suitable FE mesh construction as well as the methodology to quantify the accuracy and quality of the methods) to the physics underlying the mechanical deformation (elastic and hyperelastic parameters exposed in the literature, and loading forces and boundary condition) to solve the problem.
Our analysis begins evaluating the similarity of the glandular tissue between two mammograms from the same patient, acquired in the same day and in a short time frame. This fact allows us to evaluate the effect of the breast compression in the parenchymal pattern distribution. The monomodal analysis provides us a baseline result to perform the multimodal comparison between MRI and mammography. To achieve this goal, a fully automatic framework to register the density maps, obtained from digital mammograms and MRI images, was developed. This software uses a patient-specific biomechanical model of the breast, which mimics the mammographic compression performed during the mammographic acquisition. In this work, we propose a new methodology to project the glandular tissue directly from the MRI, avoiding the loss of information that can be yielded when the image is deformed. Our analysis shows a high similarity between the information contained in the two modalities as well as a high structural similarity in the distribution of the glandular tissue.
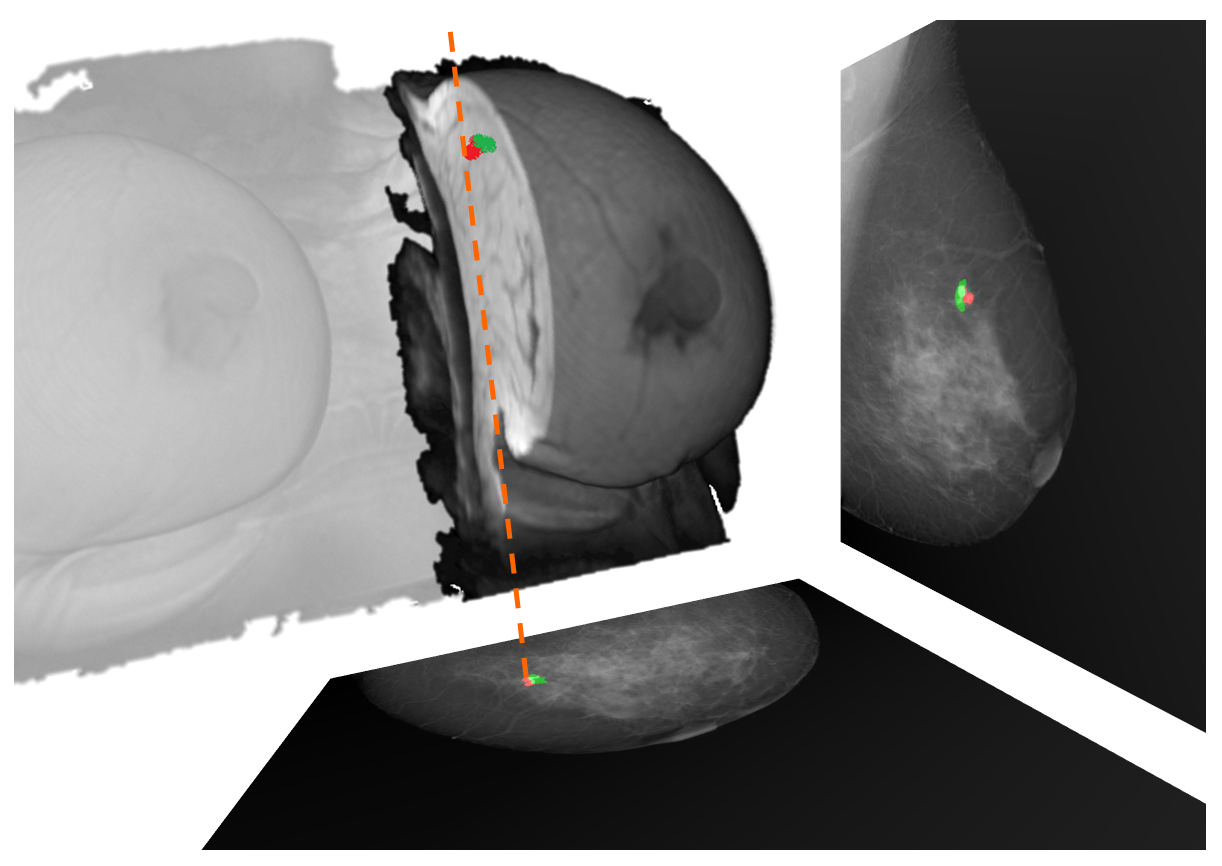
During the registration framework evaluation we computed the target registration error (TRE) between landmarks -i.e. lesions- in both MRI and mammography. The 2D problem consists of directly projecting the landmark position from the MRI to the mammogram, computing the Euclidean distance between the computed and the real landmark position. However, locating the 3D position, within the MRI, from the corresponding lesions in the mammograms, used to require complex and computational expensive methods to undo the breast compression. To solve this issue, we propose a new, fast and efficient algorithm to locate the landmark position within the MRI. Using a similar methodology to that proposed during the glandular tissue projection, a back-projection ray-tracing can be performed, allowing to locate the ray path in the uncompressed biomechanical model, avoiding to undo the breast compression. To accelerate the computation of the intersection between two rays, we propose an easy algorithm which sub-divide the rays, reducing the search space. Our model reduces the search space until 600 times with respect to a traditional point-by-point search. Thus, the computational time to locate the intersection is about 8 ms, allowing real-time applications in the clinical practice. Furthermore, the TRE, in average, are about 1 cm, better than those exposed in previous works.
To conclude, we evaluate the capability of using intensity gradient values to perform the registration between the images. In this case the glandular tissue gradient is extracted from the mammograms and MRI images by means of image processing techniques. The MRI gradient is projected to the mammogram using two different approaches. The first one uses the accumulated directional derivative, obtaining a scalar value which is comparable to the norm of the gradients obtained from the mammograms. The second one accumulates the gradient considering each direction independently, yielding a vector which is comparable to the directional derivative obtained from the mammograms by means of a gradient correlation metric. Our results show an improvement in the TRE using scalar gradient values, with respect to the traditional intensity-based approach.
In summary, this thesis will help radiologists and physicists to better understand the variations of the glandular tissue that can be clearly visible in one modality but not in the other. Furthermore, the evaluation of the information can guide researchers to obtain more accurate segmentation algorithms, considering the partial volume effect presented in the MRI, as well as to improve the multimodal image registration between the two modalities, not only by means of intensity-based methods but also considering additional information such as gradients. Our methodology includes several proposals to develop real time applications or with acceptable time values in the clinical practice.